Biodiversity science encompasses multiple disciplines and biological scales from molecules to landscapes. Nevertheless, biodiversity data are often analyzed separately with discipline-specific methodologies, constraining resulting inferences to a single scale. To overcome this, we present a topic modeling framework to analyze community composition in cross-disciplinary datasets, including those generated from metagenomics, metabolomics, field ecology and remote sensing. Using topic models, we demonstrate how community detection in different datasets can inform the conservation of interacting plants and herbivores. We show how topic models can identify members of molecular, organismal and landscape-level communities that relate to wildlife health, from gut microbes to forage quality. We conclude with a future vision for how topic modeling can be used to design cross-scale studies that promote a holistic approach to detect, monitor and manage biodiversity.
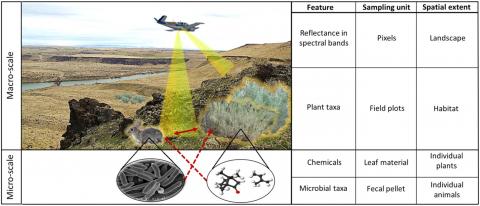
Illustration of how communities are measured in sampling units that span micro- and macro-scales. In the sagebrush steppe ecosystem, these communities are linked across scales. Microbial taxa in fecal pellets from individual herbivores interact with chemical features in leaf material when herbivores consume individual plants. Metabolite features in leaf material consumed by herbivores are dependent on the abundance of individual plant taxa detected within field plots. The distribution of plant taxa can be detected with spectral bands in pixels of aerial imagery obtained remotely within landscapes.
| GEM3 author(s) | |
| Year published |
2021
|
| Journal |
Oikos
|
| DOI/URL | |
| GEM3 component |
Mapping
Mechanisms
Modeling
|
| Mentions grant |
Yes
|